Bivariate Poisson Regression with Expectation-Maximisation
Count data is commonly modelled using a generalised linear model with a Poisson likelihood, and this approach extends trivially to cases with multiple count data random variables,
In this short post I’ll describe the case in which we have two correlated count variables and we believe that a linear function of some known covariates can explain variation in the data.
Univariate Poisson regression and the MLE fit
The poisson distribution is specified by a parameter,
Assuming the data is IID then the log-likelihood is given by:
The last piece we need, a ’link function’: There are a couple of options we can use to relate the Poisson parameter to our model parameters,
Typically we would take derivatives of the log-likelihood with respect to the
We’re nearly ready to start fitting some models, but first some housekeeping to make our life easier later:
from dataclasses import dataclass
from typing import TypeVar, Generic, Tuple, Union, Optional
import numpy as np
import pandas as pd
from scipy import optimize
@dataclass
class Dims:
N: int
M: int
@dataclass
class ObservedData:
covariates: np.array
Z0: np.array
Z1: np.array = None
@dataclass
class PoissonData:
dims: Dims
observed: ObservedData
We can generate some fake data from a true model lambdas by generating some random covariates and using some true model
def generate_poisson_data(true_betas: np.array, N: int, sigma=1) -> PoissonData:
M = true_betas.shape[0]
covariates = np.random.normal(0, sigma, (N, M))
# Fix the first row to allow for a constant term in the regression
covariates[0, :] = 1
lambdas = np.exp(covariates.dot(true_betas))
poisson_draws = np.random.poisson(lambdas)
return PoissonData(Dims(N, M), ObservedData(covariates, poisson_draws))
and, as above, in order to fit the model we need an expression for the negative (so make the problem is a minimisation) log-likelihood and for neatness, and further use, we’ll want a little wrapper around scipy’s minimise function:
def poisson_nll(betas, covariates, targets):
loglambda = covariates.dot(betas)
lambda_ = np.exp(loglambda)
return -np.sum(targets * loglambda - lambda_)
def poisson_regression(covariates, targets):
betas = np.ones(covariates.shape[1])
r = optimize.minimize(poisson_nll, betas, args=(covariates, targets), options={'gtol': 1e-2})
assert r.success, f"Poisson Regression failed with status: {r.status} ({r.message})"
return r.x
and with these we can generate, and fit, some Poisson data from the following model:
true_betas = np.array([1, 2, -1])
dataset = generate_poisson_data(true_betas, 1000)
poisson_regression(dataset.observed.covariates, dataset.observed.Z0)
>>> array([ 0.99064907, 2.00878208, -0.98579406])
Of course the usual caveat applies – if you’re trying to build a univariate Poisson regression in python, don’t use my code - do it using statsmodels:
import statsmodels.api as sm
fit = sm.GLM(
dataset.observed.Z0,
dataset.observed.covariates,
family=sm.families.Poisson()
).fit()
print(fit.summary())
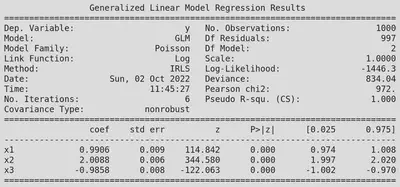
We can see that our fit is in good agreement with statsmodels, but we also get lots of other interesting information (coefficient uncertainty, NLL, deviance, etc.).
Correlations between two Poisson random variables
As mentioned in the introduction, if we have two Poisson random variables,
and to maximise this we can just maximise the two sums separately. When doing so all of derivate crossterms,
But what if we suspect that our two random variables are correlated?
The Bivariate Poisson model deals with exactly this case – let
are bivariate poisson distributed. Clearly
since sums of two independent Poisson distribution results in another Poisson parametrised by the sum of the two input distribution parameters. But more interestingly we also have
where
The PMF of the Bivariate Poisson model is given by:
We can see that when
The problem we now have to solve then is:
where
Fitting the Bivariate Poisson with the EM algorithm
We saw above that we can directly maximise the likelihood of a univariate Poisson distribution, but with the Bivariate Poisson case it is more awkward. Here I’m going to follow the approach taken in the Karlis and Ntzoufras paper 2 and take an Expectation-Maximisation approach 3 4. I also recommend Ntzoufras’s book which describes lots of interested models 5.
Computing The Likelihood
We can write down a seemingly useless for for the bivariate model, one expressed in terms of all three of the unobserved Poisson components:
We can perform a change of variables so that our model is in terms of the two observed random variables
and therefore the log-likelihood is
This is almost the same as the ‘Double Poisson’ model and we know we can maximise it with respect to the
Computing The Density of the Latent Variable
To get the
The denominator is just the Bivariate Poisson density and
and both of the distributions on the right-hand side are just more Poisson distributions. Thus:
Computing The Expectation of the Latent Variable
With this density computed we can compute the expectation of the unobserved
The denominator is independent of our index,
- Firstly we can use that fact that the
- Secondly we see that for any
Therefore
Substituting in the density of the Poisson (and suppressing, for now, the EM iteration index
We can pull out some factors of the lambdas:
Now some more index trickery, we let
we recognise the combination of factorials inside the sum as binomial coefficients
and finally we have:
The EM algorithm
The auxilliary function,
where the terms which don’t depend on
we can see that to maximise the
to compute the weights
- E-step: Using our current best guess for the model weights,
- M-step: We perform the follow three Poisson regressions
to find our next estimates for the model weights
Implementation in python
And now lets implement all of this in python. We start by generating the latent
@dataclass
class LatentData:
lambdas: np.array
Y0: np.array
Y1: np.array
Y2: np.array
@dataclass
class BivariateData:
dims: Dims
latent: LatentData
observed: ObservedData
def generate_observed_bivariate_poisson_data(YY_latent: np.array) -> np.array:
return np.stack([
YY_latent[0,:] + YY_latent[2,:],
YY_latent[1,:] + YY_latent[2,:],
])
def generate_bivariate_poisson_data(true_betas: np.array, N: int, sigma: float) -> BivariateData:
M = true_betas.shape[0]
covariates = np.random.normal(0, sigma, (N, M))
# Fix the first row to allow for a constant term in the regression
covariates[0, :] = 1
# Build the lambdas from the betas, and then sample from the Poissons
lambdas = np.exp(covariates.dot(true_betas))
latent_poisson_draws = np.random.poisson(lambdas)
observations = generate_observed_bivariate_poisson_data(latent_poisson_draws.T)
return BivariateData(
Dims(N, M),
LatentData(lambdas, *latent_poisson_draws.T),
ObservedData(covariates, *observations),
)
true_betas = np.array([
[0.3, 0.2, 0.0, 0.3, 0.0, 0.0],
[0.5, 0.0, -0.1, 0.0, 0.0, -0.5],
[0.0, 0.0, 0.0, 0.0, 1.0, 0.0],
]).T
dataset = generate_bivariate_poisson_data(true_betas, N=10000, sigma=1)
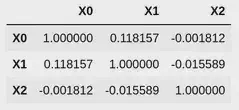
We can check the correlation matrix of the latent data, and the observed data to see that this has worked as expected:
import pandas as pd
pd.DataFrame(
data=np.array([
dataset.latent.Y0,
dataset.latent.Y1,
dataset.latent.Y2,
]).T,
columns=['Y0', 'Y1', 'Y2']
).corr()
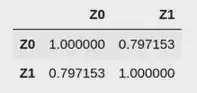
import pandas as pd
pd.DataFrame(
data=np.array([
dataset.observed.Z0,
dataset.observed.Z1,
]).T,
columns=['X0', 'X1']
).corr()
The
from scipy.special import factorial
EPS = 1e-16
def choose(n: int, p: int) -> int:
return factorial(n) / factorial(p) / factorial(n - p)
def bivariate_poisson(x: int, y: int, l0: float, l1: float, l2: float) -> float:
coeff = (
np.exp(-l0 - l1 - l2) * np.power(l0, x) *
np.power(l1, y) / factorial(x) / factorial(y)
)
p = coeff * sum([
choose(x, i) * choose(y, i) * factorial(i) * (l2 / l0 / l1) ** i
for i in range(0, min(x, y) + 1)
])
return max(p, EPS)
For the E-step of the EM algorithm we must compute the expectation
def _compute_next_y2ti(x, y, l0, l1, l2):
if min(x, y) < 1:
return 0
return l2 * bivariate_poisson(x - 1, y - 1, l0, l1, l2) / bivariate_poisson(x, y, l0, l1, l2)
def compute_next_y2ti(observed_data: ObservedData, lambdas: np.array) -> np.array:
return np.array([
_compute_next_y2ti(z0, z1, *lams)
for (z0, z1, lams) in zip(observed_data.Z0, observed_data.Z1, lambdas)
])
For the M-step we can re-use the 1D Poisson regression code from earlier in this post to compute the betas, and from the betas we get our new estimate for the lambdas:
def compute_next_betas(
data: BivariateData,
y2ti: np.array,
) -> np.array:
return np.array([
poisson_regression(data.observed.covariates, data.observed.Z0 - y2ti),
poisson_regression(data.observed.covariates, data.observed.Z1 - y2ti),
poisson_regression(data.observed.covariates, y2ti),
]).T
def compute_next_lambdas(betas, covariate_matrix):
return np.exp(covariate_matrix.dot(betas))
That is everything we really need to fit this model, but to make the output more interesting we should include a little extra code to track the negative log-likelihood of the model, and the mean squared error to see that the EM algorithm is converging:
@dataclass
class FitResult:
betas: np.array
neg_loglikelihood: float
def bivariate_poisson_nll(betas: np.array, observed_data: ObservedData):
lambdas = np.exp(observed_data.covariates.dot(betas))
return -np.sum([
np.log(bivariate_poisson(z0, z1, *lams))
for (z0, z1, lams) in zip(observed_data.Z0, observed_data.Z1, lambdas)
])
def bivariate_poisson_prediction(betas, observed_data: ObservedData):
lambdas = np.exp(observed_data.covariates.dot(betas))
return np.stack([lambdas[:, 0] + lambdas[:, 2], lambdas[:, 1] + lambdas[:, 2]])
def compute_mse(predictions, Z0, Z1):
return np.mean((predictions[0,:] - Z0 + predictions[1,:] - Z1) ** 2)
def compute_bivariate_poisson_data_mse(betas: np.array, observed_data: ObservedData) -> float:
preds = bivariate_poisson_prediction(betas, observed_data)
return compute_mse(preds, observed_data.Z0, observed_data.Z1)
And finally we fit the model:
def bivariate_poisson_fit_using_em(
dataset: BivariateData,
em_max_itr: int=20,
logging: bool=False
) -> FitResult:
# Construct some initial guesses for the lambdas and betas
betas = np.zeros((dataset.dims.M, 3))
lambdas = compute_next_lambdas(betas, dataset.observed.covariates)
for itr in range(1, em_max_itr + 1):
nll = bivariate_poisson_nll(betas, dataset.observed)
bivpoisson_mse = compute_bivariate_poisson_data_mse(betas, dataset.observed)
if logging:
print(f"{itr:<4} {nll:<10.3f} {bivpoisson_mse:.3f}")
# E-step:
y2ti = compute_next_y2ti(dataset.observed, lambdas)
# M-step:
betas = compute_next_betas(dataset, y2ti)
lambdas = compute_next_lambdas(betas, dataset.observed.covariates)
return FitResult(betas, nll)
fit_bp = bivariate_poisson_fit_using_em(dataset, provide_model_structure=False, logging=True)
>>> 1 45134.088 33.348
>>> 2 34808.743 11.267
>>> 3 34207.242 10.415
>>> 4 33747.553 9.645
>>> 5 33498.612 9.208
>>> 6 33406.624 9.064
>>> 7 33381.469 9.042
>>> 8 33375.651 9.046
>>> 9 33374.371 9.052
>>> 10 33374.077 9.054
we can compare the model fitted
fit_bp.betas.round(2).T
>>> array([[ 0.3 , 0.21, -0. , 0.3 , 0. , 0.02],
>>> [ 0.5 , -0. , -0.1 , -0.01, -0.01, -0.5 ],
>>> [-0. , -0.01, -0.01, -0.01, 1. , 0. ]])
true_betas.round(2).T
>>> array([[ 0.3, 0.2, 0. , 0.3, 0. , 0. ],
>>> [ 0.5, 0. , -0.1, 0. , 0. , -0.5],
>>> [ 0. , 0. , 0. , 0. , 1. , 0. ]])
We can also perform a Double Poisson fit and compare the MSE of the two models.
def double_poisson_fit(data: BivariateData) -> FitResult:
betas = np.array([
poisson_regression(data.observed.covariates, data.observed.XX),
poisson_regression(data.observed.covariates, data.observed.YY),
]).T
return FitResult(betas)
def double_poisson_prediction(betas, data: ObservedData):
lambdas = np.exp(data.covariates.dot(betas))
return np.stack([lambdas[:, 0], lambdas[:, 1]])
fit_dp = double_poisson_fit(dataset)
preds_dp = double_poisson_prediction(fit_dp.betas, dataset.observed)
print(compute_mse(preds_dp, dataset.observed.XX, dataset.observed.YY))
>>> 16.204314367381187
which is significantly worse than the 9.054 managed by the Bivariate Poisson regression.