The EM Algorithm Part 1: Gaussian Mixture Models
When given the task of estimating the density of some simple dataset with a unique mode we can choose a family of distributions (e.g. Normal, t-distribution, etc.) and use Maximum Likelihood Estimation to find the parameters which best describe our data. But when the dataset has a more complex multimodal structure, we might instead want to model the data as a latent mixture model.
In a latent mixture model we posit that each observed datapoint is generated by a combination of several random variables; usually a known and fixed number of random variables from the same family. The samples from each base random variable is hidden from us as we only observe their combined effect, as a result we cannot use Maximum Likelihood Estimation to fit the data directly and must apply a technique known as Expectation-Maximisation.
Here, we’ll first look at fitting a simple dataset by directly maximising the likelihood function before deriving the Expectation-Maximisation algorithm and using it to fit a Gaussian Mixture Model to a more complicated fake dataset.
I recommend the following text books on this subject 1 2 3 4.
Maximum Likelihood Estimation of a simple dataset
Let’s jump straight in with some code. If we want to estimate the density of some one dimensional data,
example_data = stats.t(5, 10, 5).rvs(400)
t_args = stats.t.fit(example_data)
n_args = stats.norm.fit(example_data)
calculate_ll = lambda model, args, data: np.sum([model(*args).logpdf(x) for x in data])
n_ll = calculate_ll(stats.norm, n_args, example_data)
t_ll = calculate_ll(stats.t, t_args, example_data)
print(n_ll, t_ll)
>>> -1237.30, -1260.66
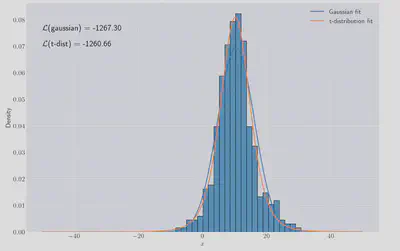
Both models visually like they fit OK but the
If we dig through the scipy documentation 5 we’ll find that continuous random variable like the two models we’re looking at are fitted exactly by finding the MLE values. For certain distributions, like Gaussians, we can easily derive the values of the MLE parameters so let’s do it.
For this model we have the following expression for the loglikelihood
which we can differentiate with respect to
I.e. the MLE estimate for the mean parameter in a normal distribution fit is just the empirical mean. Proceeding similarly, for
which is the empirical standard deviation. We can confirm this is true for the dataset we have here:
print(example_data.mean(), example_data.std())
print(*n_args)
>>> 10.051125578381319 6.149058201486978
>>> 10.051125578381319 6.149058201486978
So far so good! We could do something similar for the
Gaussian Mixture Models
Things get more complicated when our model involves ’latent’ variables. Latent variables are ones for whom we never get to observe, the best we can do is to try to estimate their values. The canonical example of a latent variable model is the Gaussian Mixture Model (GMM) which we’ll go through in detail here.
Let’s build another dataset which we can then try and fit a GMM model to:
true_params = ((-5, 3), (-1, 1), (3, 1))
N, M = 999, 3
YY = [
stats.norm(mu, sigma).rvs(N // M) for (mu, sigma) in true_params
]
and plotting that data:
_, ax = plt.subplots(figsize=(18, 7), ncols=2)
sns.histplot(YY, bins=50, ax=ax[0])
# 'Anonymise' the data
YY = np.concatenate(YY)
sns.histplot(YY, bins=50, ax=ax[1], color='b');
[a.set(xlabel=r'$x$') for a in ax];
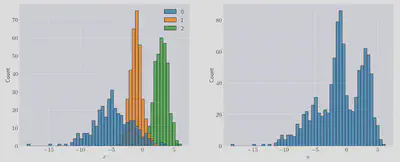
In the left-hand panel we can see the composite distributions which make up the resulting mixture, shown on the right-hand panel.
We’re going to posit that this data was generated a sum of gaussians:
The problem is we don’t know
- The parameters of these Gaussians,
- Which gaussian was responsible for generating each point.
The last point, (3), is really the crux of the problem since if we knew which gaussian each point was derived from then we do exactly what we did above - use a fitting procedure to calculate the mean and variance parameters and we’d be done.
This uncertainty is exactly what our latent variables encode. Let’s say we propose (correctly!) that there are three gaussians in our mixture,
Formally, we’re proposing that
We can write down the likelihood for this model but because we don’t observe the
The EM Algorithm
Let’s derive the EM algorithm before we apply it to the GMM problem above. We start, as before, by writing down the loglikelihood over the observed data.
We now need to find a way to introduce our
The law of conditional probability allows us rewrite the term inside the logarithm as follows
So, we have broken the incomplete log-likelihood into two terms:
- The ‘auxilliary’ function,
- An ’entropy’ term,
We have all the ingredients to compute
where we have simply multiplied and divided by
We can now apply Jensen’s inequality for concave functions
to swap the expectation and the logarithm and get a lower bound on the loglikelihood:
Now define
And, going step-by-step for clarity, we take
and by the construction of
but the right-hand side we can identify as equation
which proves that maximising
There is a lot going on here so lets break it down. We’ve found a function,
The schematic figure below illustrates this process. We start with some guess
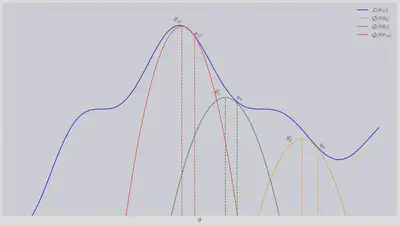
In summary, the EM algorithm breaks down into two steps:
- E-step: Given values for
- M-step: Maximise
We repeat these steps until we’re happy it has converged (using either the log-likelihood of convergence of the parameters,
Fitting Gaussian Mixture Models using EM
Let’s apply all this to the Gaussian Mixture Model. As a reminder, the full model is:
where
The E-step demands that we compute the auxilliary function:
where we have writen the categorical distribution as a product using an indicator variable.
The term in the log is constant with respect to the expectation so we need only the expectation of the indicator which is given by
therefore
We can calculate the posterior over the
Therefore,
That’s the E-step done. The M-step requires us maximise this expression to get an update rule for the model parameters. I’ll demonstrate this is only for
and the constraint over
The other update rules are:
and that condlues the M-step. We can now implement this and see if we can fit the dataset.
Implementing EM for GMMs in python
from typing import Tuple
def get_initial_guess_for_mus(YY: np.array, n_components: int) -> np.array:
# Choose some sensible percentiles
return np.percentile(YY, np.linspace(0, 100, n_components + 2)[1: -1])
def compute_q(w_ik, phis, mus, sigmas, YY):
N, n_components = w_ik.shape
return np.sum([
w_ik[i, k] * (np.log(phis[k]) + stats.norm(mus[k], np.sqrt(sigmas[k])).logpdf(YY[i]))
for i in range(N)
for k in range(n_components)
])
def compute_h(w_ik):
N, n_components = w_ik.shape
return - np.sum([
w_ik[i, k] * np.log(w_ik[i, k])
for i in range(N)
for k in range(n_components)
])
def fit_gmm_with_em(YY: np.array, n_components: int, em_itrs: int=10) -> Tuple[np.array]:
N = YY.shape[0]
phis = np.ones(n_components) / n_components
range_ = YY.max() - YY.min()
sigmas = np.ones(n_components)
w_ik = np.zeros((N, n_components))
mus = get_initial_guess_for_mus(YY, n_components)
for itr in range(em_itrs):
# E-step
w_ik = np.array([
phis[k] * stats.norm(mus[k], np.sqrt(sigmas[k])).pdf(YY[i])
for i in range(N)
for k in range(n_components)
]).reshape(N, n_components)
w_ik = w_ik / w_ik.sum(axis=1)[:, np.newaxis]
# M-step
sum_i_w_ik = w_ik.sum(axis=0)
phis = sum_i_w_ik / N
mus = (w_ik.T.dot(YY)) / sum_i_w_ik
sigmas = w_ik.T.dot((YY[:, np.newaxis] - mus) ** 2).diagonal() / sum_i_w_ik
# Compute the value of the Q function at our current parameters
print(itr, compute_q(w_ik, phis, mus, sigmas, YY))
return w_ik, phis, mus, sigmas
w_ik, phis, mus, sigmas = fit_gmm_with_em(YY, 3)
We can plot the individual gaussians as well as the resulting mixture to check the fit:
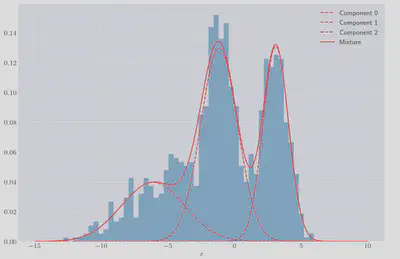
It looks like a much better fit than if we’d used a single gaussian! We can also calculate the log-likelihood of the GMM fit, to what we can achieve if we just fit a single Gaussian to the data:
# Our proxy objective function, Q
Q = compute_q(w_ik, phis, mus, sigmas, YY)
# The entropy term, H
H = compute_h(w_ik)
single_gaussian_ll = calculate_ll(stats.norm, n_args, xx)
print(Q + H, single_gaussian_ll)
>>> -2586.743170746824, -2748.671049075651
Unsuprisingly the mixture model gives a much better fit!
Closing Remarks
We’ve derived and then applied the Expectation-Maximisation algorithm to some simple fake one-dimensional data and shown that it outperformed the naive single gaussian fit by checking the log-likelihood of the two approaches.
In part 2 of this blog I’ll take the theory explained here and look at a very different application: How we can deal with ‘censored data’ problems (where data is, for whatever reason, only partially observed) using expectation maximisation.